In eukaryotes, the ubiquitin (Ub) system has emerged at the center of many signaling mechanisms through their targeting of specific key proteins for ubiquitination and degradation by the 26S proteasome. The specificity of ubiquitination is conferred by Ub E3 ligase complexes, which serves as the substrate receptor and recruits E2 enzymes that catalyze transfer of Ub to a substrate lysine. Typically, E3s recognize substrates that are modified, often by phosphorylation (for more details, read Zheng & Shabek and Shabek & Zheng and Shabek & Ciechanover). Remarkably, plants evolved the Ub system to control numerous growth and developmental processes including senescence, embryogenesis, defense, hormone signaling, and environmental responses. Plants genome predicts more than 1,400 potential Ub ligases, almost twice higher number that most of them are without obvious counterparts in other eukaryotes. 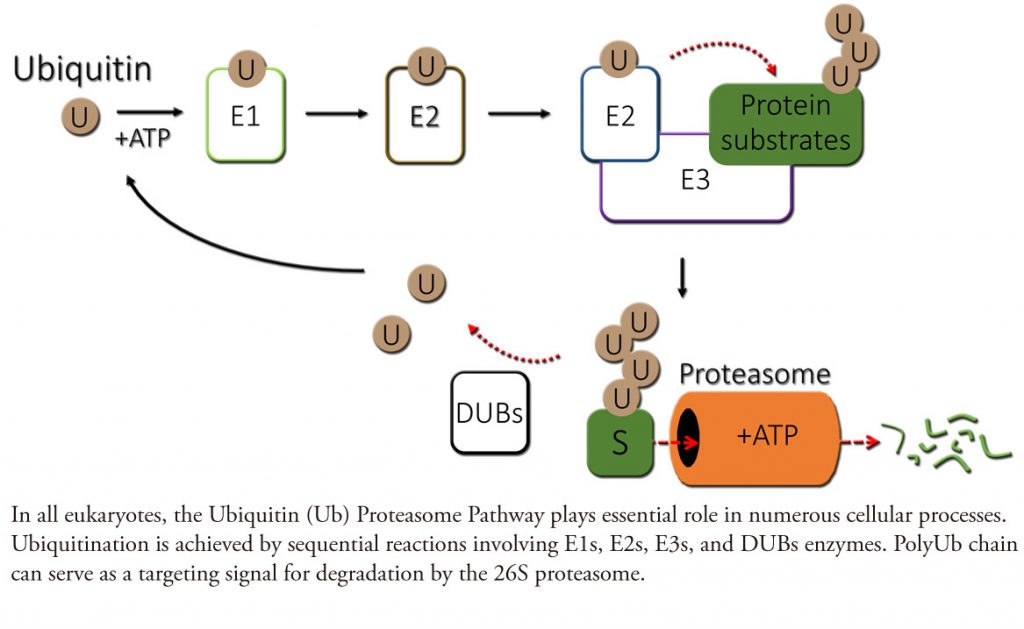
Despite the significant advances made in understanding the cellular mechanisms that underlie signal-dependent substrate recognition by the Ub system in plants, this field is largely unexplored. This notion is especially significant given the number of uncharacterized E3 ligases, target substrates, and countless signals that are yet to be unveiled. Our research aims to reveal and dissect how E3 ligases regulate cellular response to a variety of stimuli, and what the roles are of other components of the Ub system, including deubiquitinating enzymes, and Ub-like proteins in plants.
For our research on the roles of E3 ubiquitin ligases in hormone signaling, please go to Hormone Sensing & Signaling page where you will find more information about the lab work on the E3 MAX2/D3 that regulates Strigolactones (SL) as well as the Karrikin (KAI2-mediated) signaling pathway.
In the lab we are interested in uncovering the proteome landscape of E3 ligases in planta and their entire interactomes (omic profiling of interacting proteins). Together with collaborators, we have developed and optimized advanced proximity labeling strategies to identify in vivo interactomes. We have recently showcased our first sets of interactomes by focusing on the SCF E3 ubiqutin ligase (published in New Phytologist Sun et al., 2024):
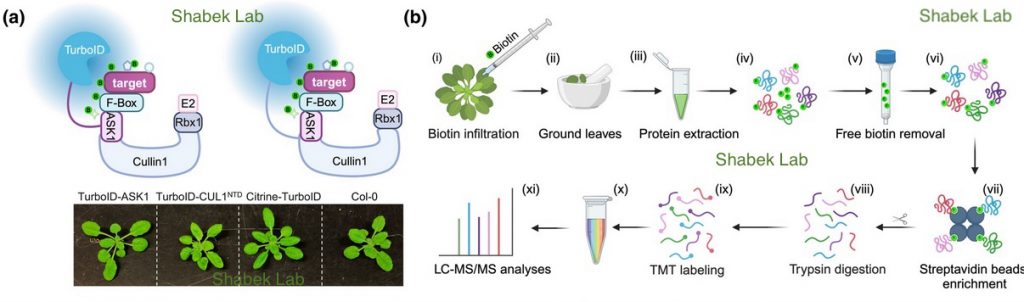
 The S-phase Kinase-Associated Protein 1 (SKP1, ASK1 in Arabidopsis) and Cullin1 are core components of the SCF complex (SKP1, Cullin1, F-box protein), an E3 ubiquitin ligase that polyubiquitinates a vast array of protein targets, marking them for proteasomal degradation. Arabidopsis thaliana is predicted to have hundreds of E3s and numerous targets. However, identifying ubiquitin system targets has been challenging due to their relatively weak/transient interactions with E3 complexes and rapid degradation. We used Arabidopsis transgenic lines expressing ASK1 or Cullin1 (CUL1) fused to TurboID and performed proximity labeling proteomics to identify interacting targets.
The S-phase Kinase-Associated Protein 1 (SKP1, ASK1 in Arabidopsis) and Cullin1 are core components of the SCF complex (SKP1, Cullin1, F-box protein), an E3 ubiquitin ligase that polyubiquitinates a vast array of protein targets, marking them for proteasomal degradation. Arabidopsis thaliana is predicted to have hundreds of E3s and numerous targets. However, identifying ubiquitin system targets has been challenging due to their relatively weak/transient interactions with E3 complexes and rapid degradation. We used Arabidopsis transgenic lines expressing ASK1 or Cullin1 (CUL1) fused to TurboID and performed proximity labeling proteomics to identify interacting targets. 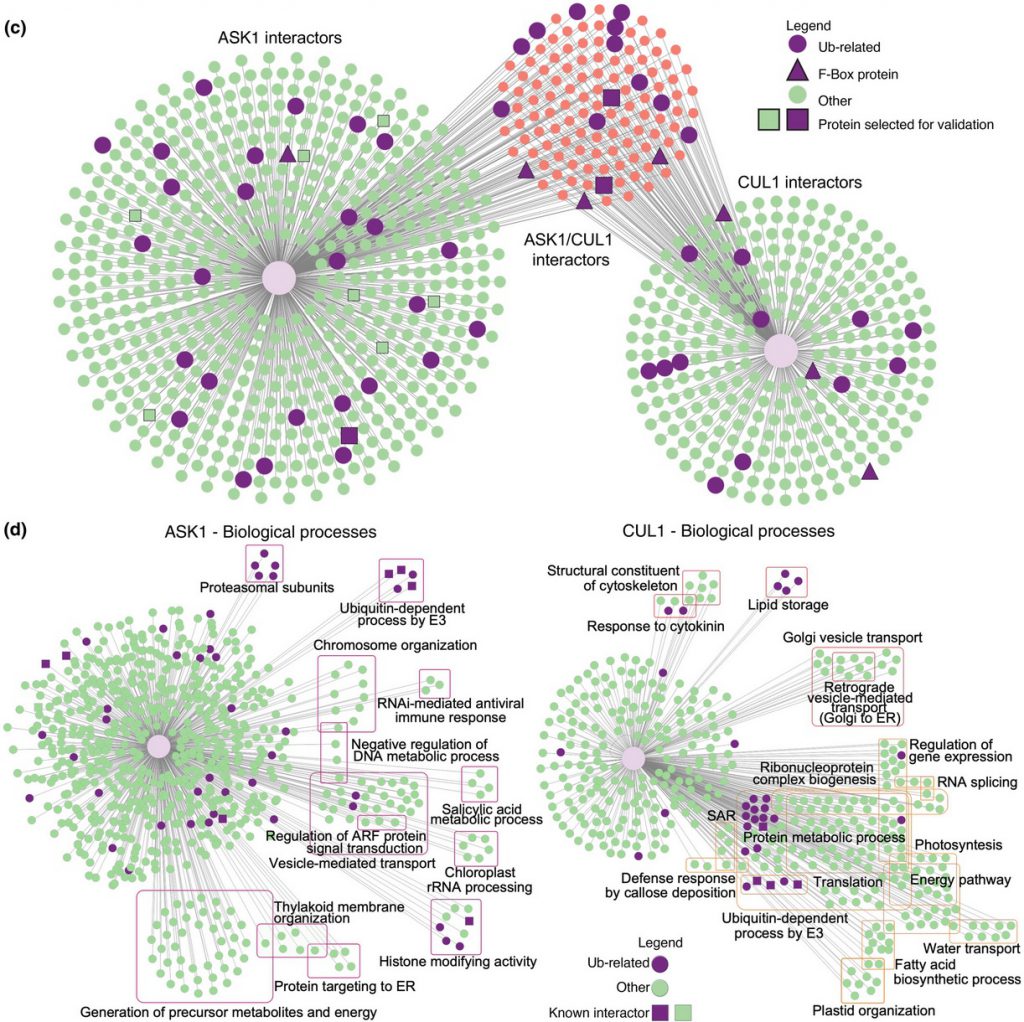
The landscape of the CUL1/ASK1 proxitome presented here provides a valuable resource for enhancing our comprehension of the biological roles of interactors and targets involved in SCF-mediated cellular functions in plants. TurboID-based PL strategy outlined in this study has the potential to be expanded to other E3 families, thereby broadening our understanding of the ubiquitin system’s repertoire under both normal and stressed environmental conditions.
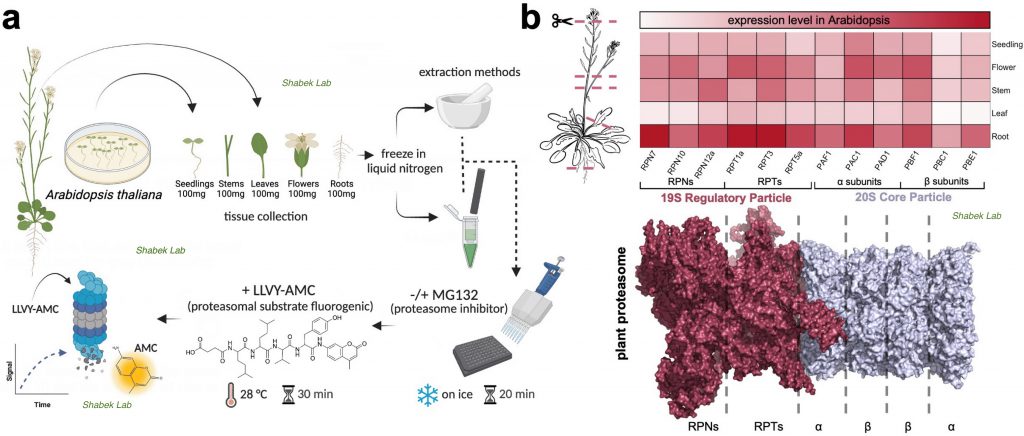
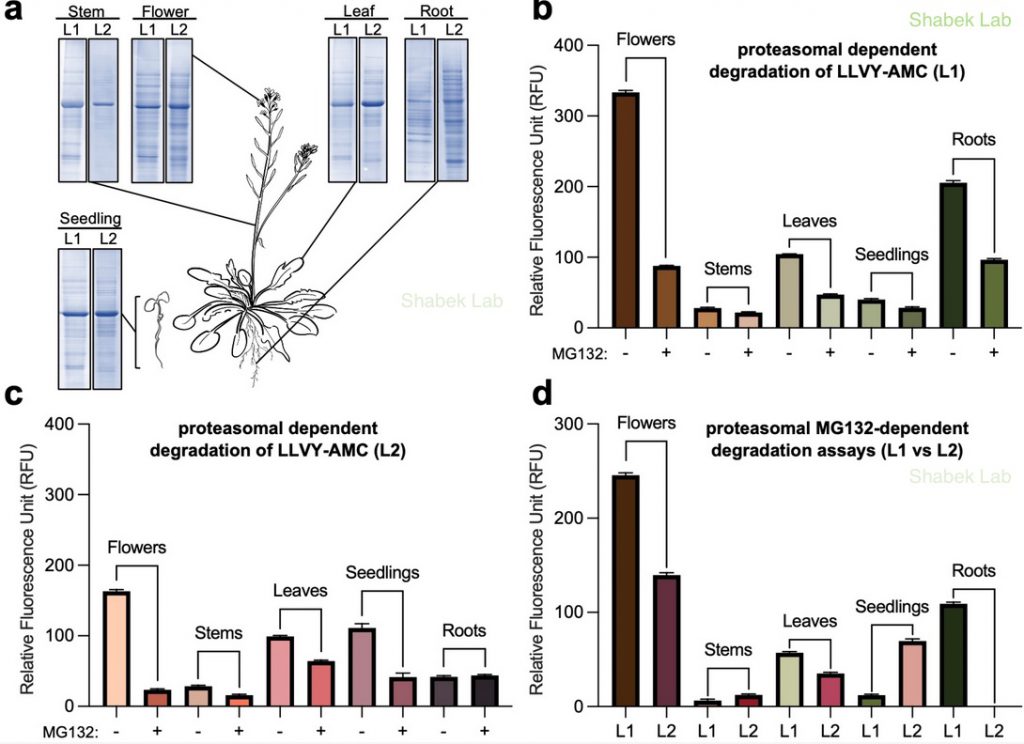
We have also recently examined the activity of 26S proteasome in distinct plant tissues using different extraction procedures. The proteasome is a massive (2.5 MDa) multi-catalytic protease complex where ubiquitinated targeted proteins are degraded through an ATP-dependent proteolysis process. In a study published in Plants (Jagadeesan, Hand, and Shabek. 2024), we utilize a fluorogenic reporting assay using two extraction methods to survey proteasomal activity in different Arabidopsis thaliana tissues. This study provides new insights into the enrichment of activity and varied presence of proteasomes in specific plant tissues.