Hormonal cues regulate many aspects of plant growth and development, as well as maintain the ability to systemically respond to environmental changes. Elucidating the molecular mechanisms governing these signaling pathways is crucial to the understanding of how plants function. Structural and functional biology have been essential keys to decode plant genetic findings and reveal the precise molecular action at the protein level. Plant hormones (aka phytohormones) comprise a set of structurally unrelated small organic compounds. Interestingly, most phytohormone signaling pathways are tightly regulated by highly coordinated intracellular protein degradation machine known as the ubiquitin-proteasome system (UPS). The specificity of UPS is conferred by the action of a family of E3 ubiquitin ligase enzymes that target specific proteins for destruction in a timely manner. Read our lab’s review (Tal et al, 2020. Plant Physiology), and a short review (Shabek et al., 2014. Nature Structural Molecular biology) for more information.
The Shabek lab is particularly interested in a novel class of plant hormones, Strigolactones (SL) as well as the Karrikin (KAI2-mediated) signaling pathway. Read our lab’s review on Strigolactones: diversity, perception, and hydrolysis (Guercio et al 2023). SLs regulate distinct aspects of plant development, promote symbiotic associations with arbuscular mycorrhizal fungi, and trigger germination of parasitic weeds. The central questions concerning the exact molecular mechanisms of SL perception and signal transduction remain to be answered and our long-term research goal is to determine how SLs control these processes at the molecular level. SLs’ perception and signal propagation are coordinated by three evolutionarily conserved proteins: the ubiquitin ligase MAX2, the SL receptor and hydrolase D14, and the novel AAA+ ATPase transcriptional repressors SMXL6/7/8 (SMXLs). Loss of function mutations in these genes result in SL-insensitive plants with numerous developmental defects including enhanced seed dormancy, reduced seedling photomorphogenesis, increased axillary branching, reduced plant height, delayed leaf senescence, and altered leaf and root development.
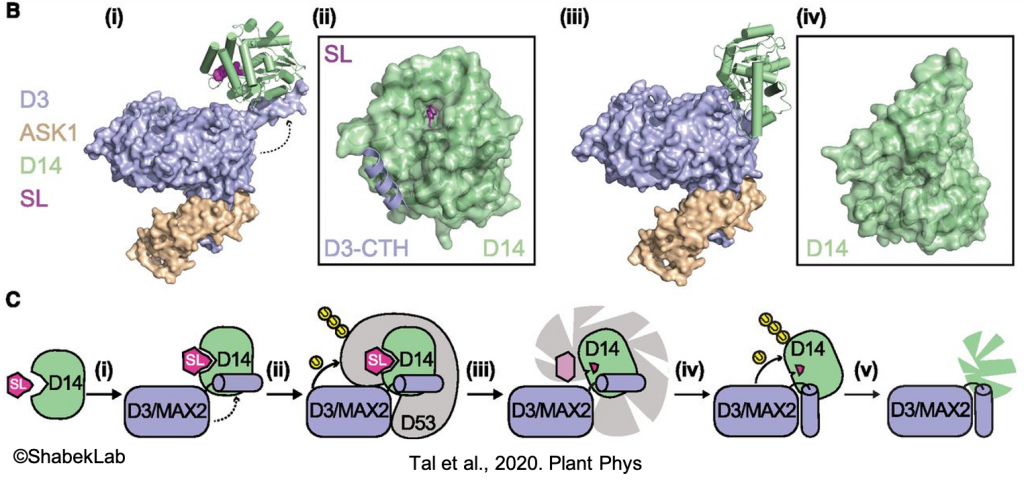
The current model suggests that the MAX2-D14 complex mediates SL response by ubiquitinating and degrading SMXLs, thus alleviating repression of SL-signaling genes. In one of our studies (published in Nature), we isolated the MAX2-D14-SL complex and determined multiple functional conformational states. These findings now open up new avenues of research and applications.
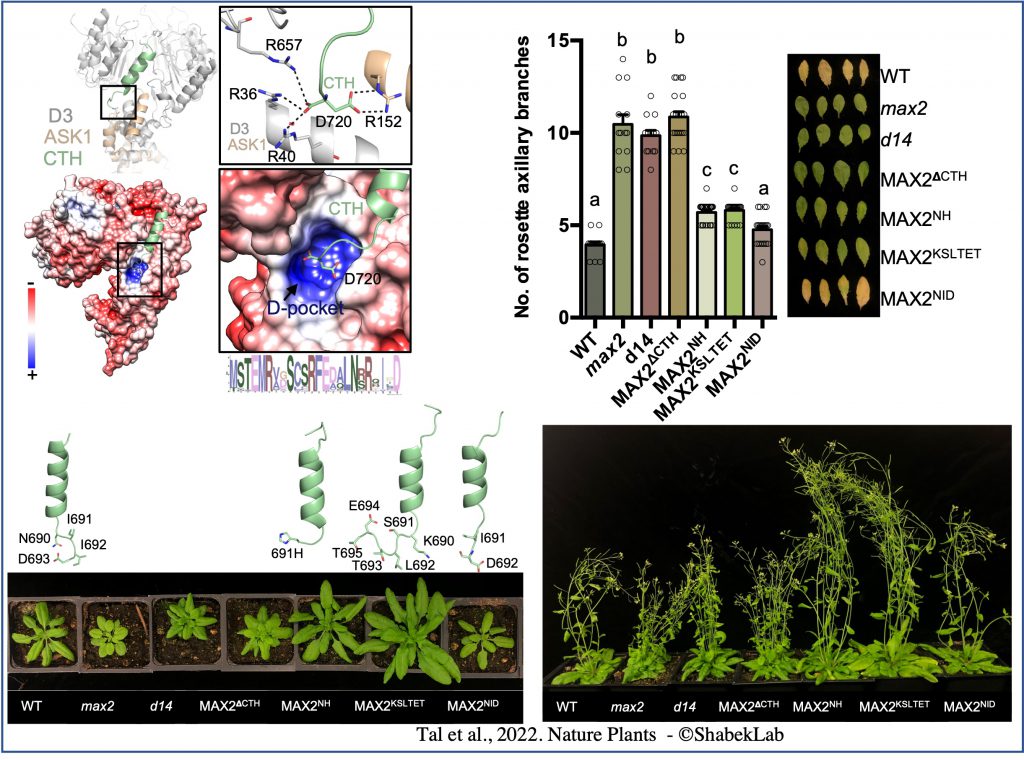 In our work (published in Nature Plants) we elucidate the functional dynamics of ASK1–D3/MAX2 in SL signalling by leveraging conformational switch mutants both in vitro and in plants. We determine the crystal structure of a dislodged D3/MAX2 mutant and demonstrate that disruptions in CTH plasticity via either CRISPR–Cas9 genome editing or expression of point mutation mutants result in impairment of SL signalling. We show that the conformational switch in ASK1–D3/MAX2 CTH directly regulates ubiquitin-mediated protein degradation. Interstingly, we uncovered an organic acid metabolite that can directly trigger the conformational switch.
In our work (published in Nature Plants) we elucidate the functional dynamics of ASK1–D3/MAX2 in SL signalling by leveraging conformational switch mutants both in vitro and in plants. We determine the crystal structure of a dislodged D3/MAX2 mutant and demonstrate that disruptions in CTH plasticity via either CRISPR–Cas9 genome editing or expression of point mutation mutants result in impairment of SL signalling. We show that the conformational switch in ASK1–D3/MAX2 CTH directly regulates ubiquitin-mediated protein degradation. Interstingly, we uncovered an organic acid metabolite that can directly trigger the conformational switch.
KAI2-mediated signaling pathway. Karrikins (KARs) are a family of butenolide small molecules produced from the combustion of vegetation and are bio-active components of smoke. These molecules are capable of inducing germination of numerous plant species, even those not associated with fire or fire-prone environments. There are striking similarities between KAR and strigolactone (SL) signaling pathways as both dependent on similar key proteins: a KAR receptor α/β hydrolase KARRIKIN INSENSITIVE2 (KAI2), MAX2 ubiquitin ligase, and the target of ubiquitination and degradation, the transcriptional corepressor SMAX1/SMXL2. KAR signaling is important in the regulation of a number of plant developmental processes including seedling development, leaf shape, cuticle formation, and root development . Furthermore, they play critical roles in arbuscular mycorrhiza (AM) symbiosis and abiotic stress response. Our lab investigates the karrikin/KAI2-ligand (KL) mediated signaling using multiple strategies in including structural biology, biochemistry and plant biology.
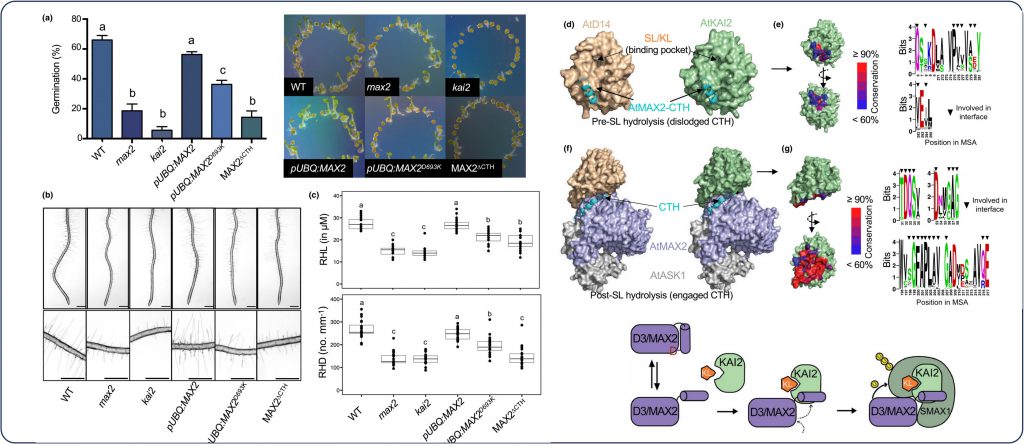
Given the dual role of the ubiquitin ligase F-box D3/MAX2 in SL and KAR/KL signaling pathways, the function of the CTH plasticity in KAR/KL signaling regulation remains to be addressed. In our study (published in New Phytologist), we investigated the function of MAX2-CTH dynamics both in vitro and in planta and found that the central role of CTH is conserved between SL and KAR/KL signaling pathways.
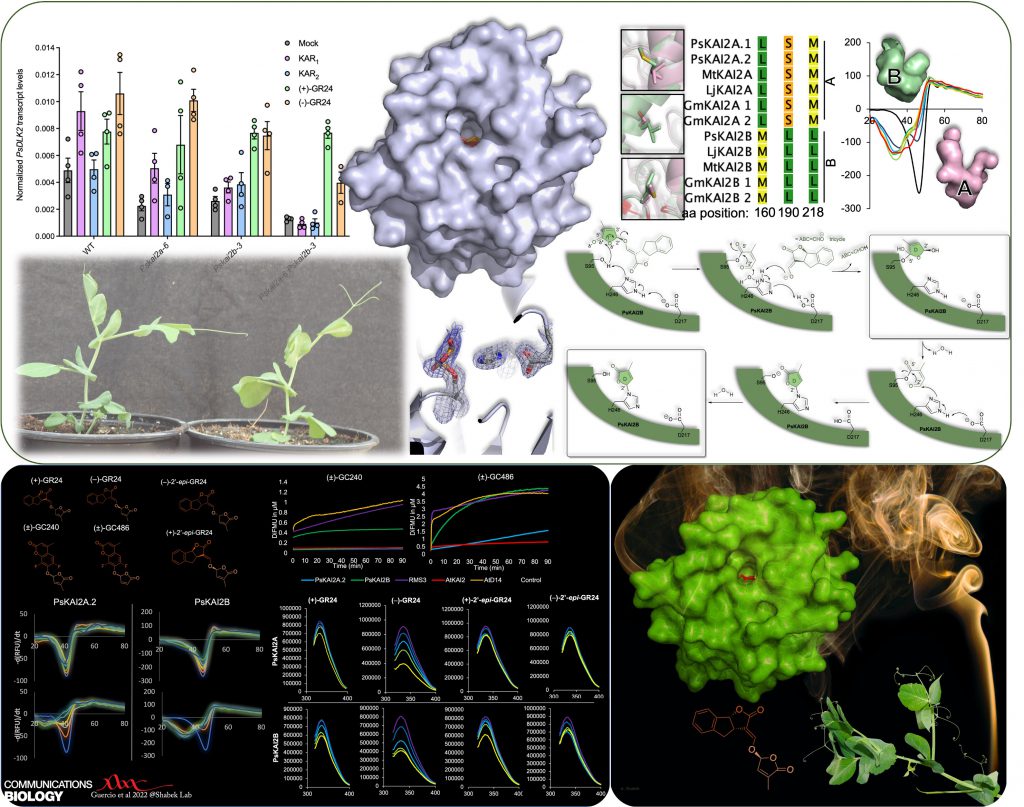
In another study (published in Communications Biology -Nature), in collaboration with multiple groups, we identified and characterized divergent KAI2 receptors in legume (pisum/pea), revealed their distinct function and ligand sensitivities, and illuminated the KAI2s enzymatic mechanism. Our research shed light on the evolution of plant hydrolase receptors and their functional adaptation in KAR/KL/SL sensing, in particular in a key crop.
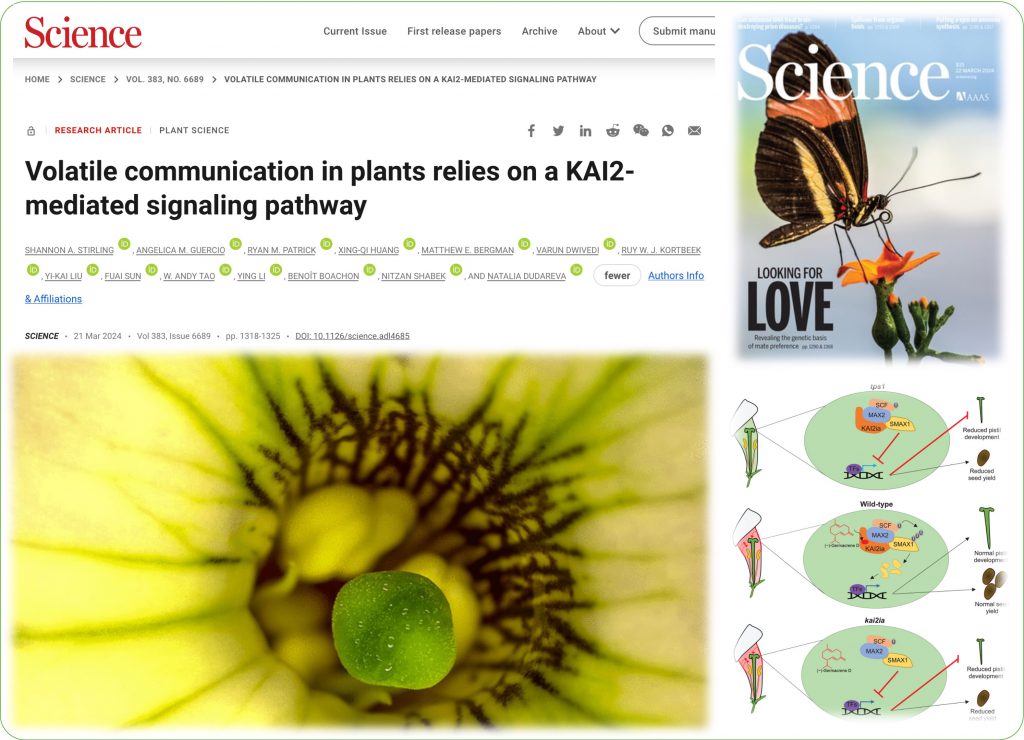 In another study (published in SCIENCE )on KAI2-mediated signaling was a product of a fruitful collaboration with Natalia Dudareva’s lab at Purdue University. Our groups are also funded by National Science Foundation (NSF-IOS collaborative grant)
In another study (published in SCIENCE )on KAI2-mediated signaling was a product of a fruitful collaboration with Natalia Dudareva’s lab at Purdue University. Our groups are also funded by National Science Foundation (NSF-IOS collaborative grant)
Plant responses to chemical signals carried through the air are critical for development, fitness, and adaptation. Stirling et al. identified a mechanism for the perception of the volatile compound (−)-germacrene D through a homolog of the Karrikin receptor KAI2 in petunia pistils. Genetic and biochemical experiments linked this perception with downstream signaling proteins and transcriptional targets. Perturbing the function of some of these components affected volatile-mediated communication and plant fitness.
In rice, several research groups, have established the physiological roles of KAI2 (D14-like) in the colonization of rice roots and the formation of symbiotic interactions with arbuscular mycorrhizal fungi (AMF). Despite the growing number of studies related to KAI2’s physiological roles, the structure and function of the rice KAI2 receptor remain unresolved, as well as its interactions with other key components of the signaling pathway in rice. 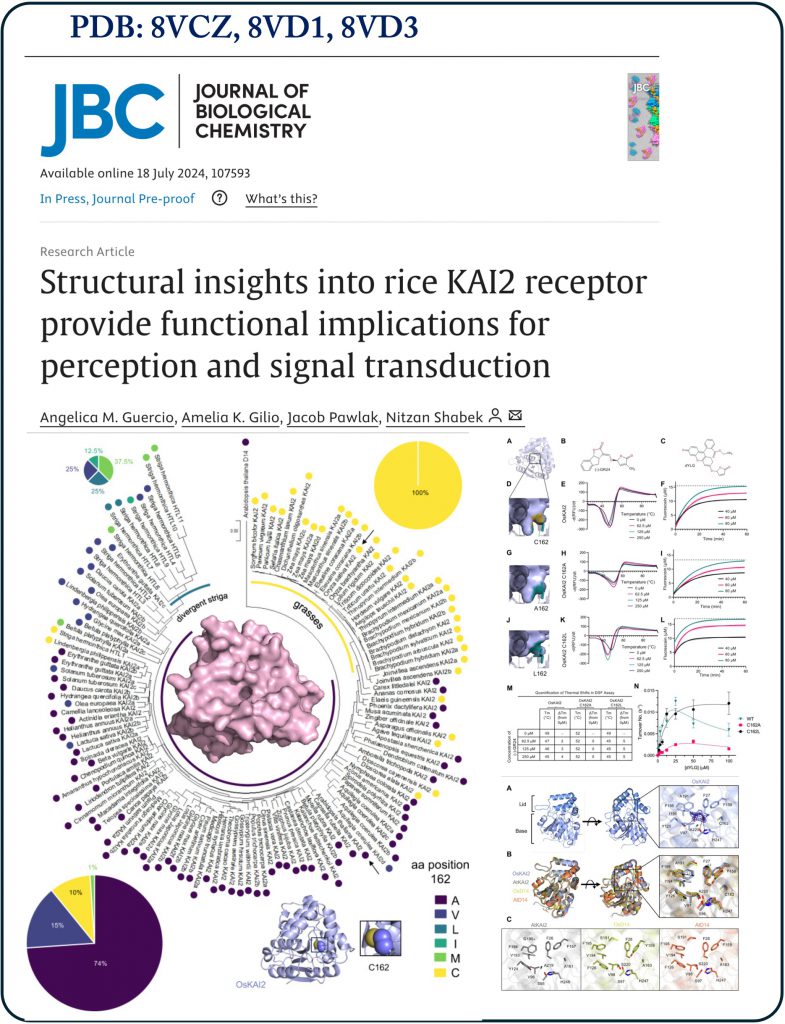 In a study we published in the Journal of Biological Chemistry (JBC) we provide structural and biochemical examinations of the rice KAI2-mediated signaling receptor hydrolase, KAI2. We have determined the first crystal structure of the receptor hydrolase and studied its sensitivity to enantiomer of 5-deoxystrigol (mimic KAI2-ligand). Using biochemical and structurally guided approaches, we reveal distinct yet highly conserved residues that play a role in ligand selectivity and affinity, as well as in binding the signal transduction components MAX2/D3 (ubiquitin ligase) and SMAX1 (transcriptional co-repressor) through binding assays and rice cell-free degradation system. To better characterize rice KAI2, we have determined the crystal structure of Arabidopsis D14 (which can also perceive strigolactones) in higher resolution then previously reported and provided a comparative analysis at the atomic level between KAI2s and D14s focusing on their catalytic pocket.
In a study we published in the Journal of Biological Chemistry (JBC) we provide structural and biochemical examinations of the rice KAI2-mediated signaling receptor hydrolase, KAI2. We have determined the first crystal structure of the receptor hydrolase and studied its sensitivity to enantiomer of 5-deoxystrigol (mimic KAI2-ligand). Using biochemical and structurally guided approaches, we reveal distinct yet highly conserved residues that play a role in ligand selectivity and affinity, as well as in binding the signal transduction components MAX2/D3 (ubiquitin ligase) and SMAX1 (transcriptional co-repressor) through binding assays and rice cell-free degradation system. To better characterize rice KAI2, we have determined the crystal structure of Arabidopsis D14 (which can also perceive strigolactones) in higher resolution then previously reported and provided a comparative analysis at the atomic level between KAI2s and D14s focusing on their catalytic pocket.
Plant Hormones Catabolism
Signaling hormones are crucial for regulating plant growth and development, with their levels meticulously controlled by biosynthesis and degradation rates. While hormone synthesis has been extensively studied, the pathways for their degradation have remained largely unexplored. For example, the synthesis pathway of strigolactones in plants is well understood, the mechanism for their breakdown however had remained elusive until recently. A newly discovered enzyme, carboxylesterase, proposed to hydrolyze and thus destroy strigolactones, has been identified as a key player in controlling strigolactone activity in plants.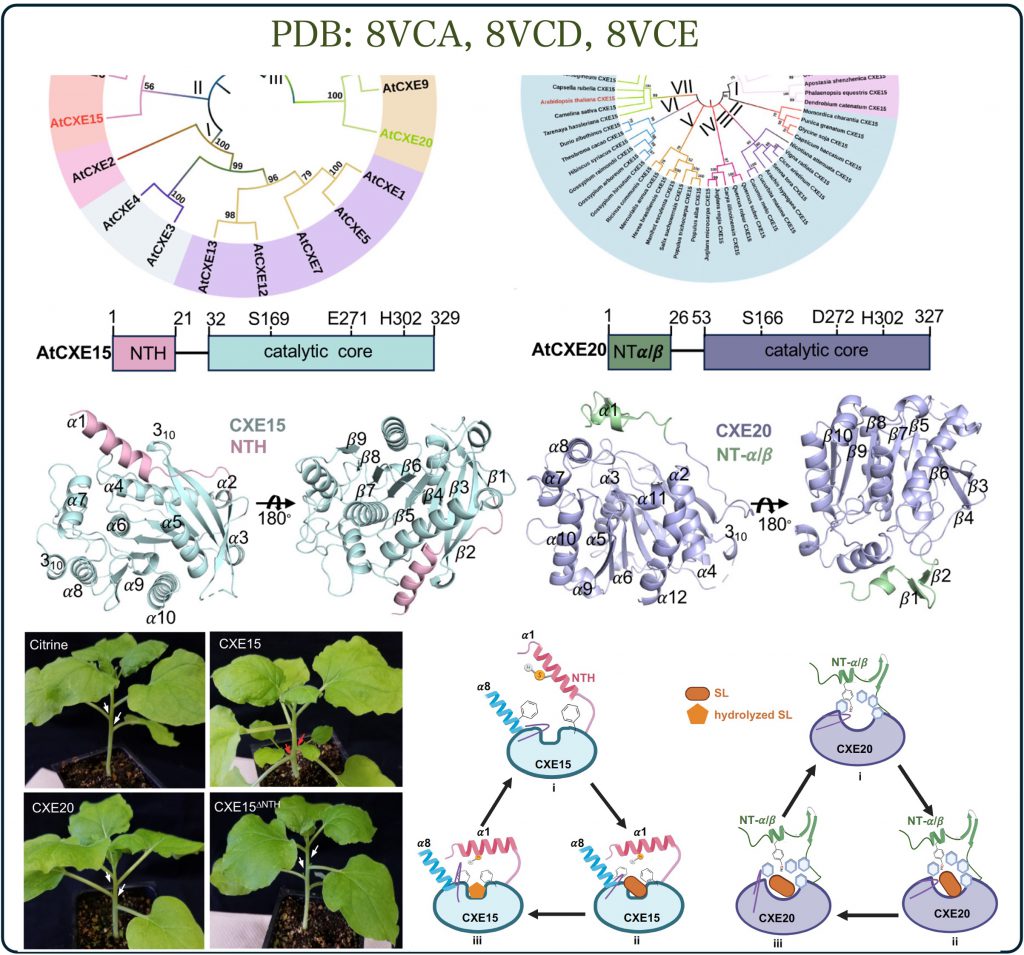

The Shabek lab, in collaboration with other experts, employed structural biology, biochemistry, and plant biology techniques to uncover how these enzymes function. Our research (published in Nature Communications, Palayam et al., 2024) reveals an unexpected mode of action, offering insights not only into strigolactone hormones but also into the mechanisms of highly conserved enzymes across different life forms.
