Dissecting Molecular Mechanisms of Signaling Pathways in Plants and their Regulation by Proteolytic System
All kingdoms of life have evolved to sense signals such as light, gravity, temperature, oxygen, metabolites, and hormones. Once perceived, these signals are transduced into developmental and physiological responses by reprogramming appropriate genes. We aim to decode and provide mechanistic insight into the basis of signal recognition and propagation pathways in cells. Our research has been focused on the investigation of the ubiquitin proteasome system, light signaling pathways, and plant hormone sensing mechanisms.
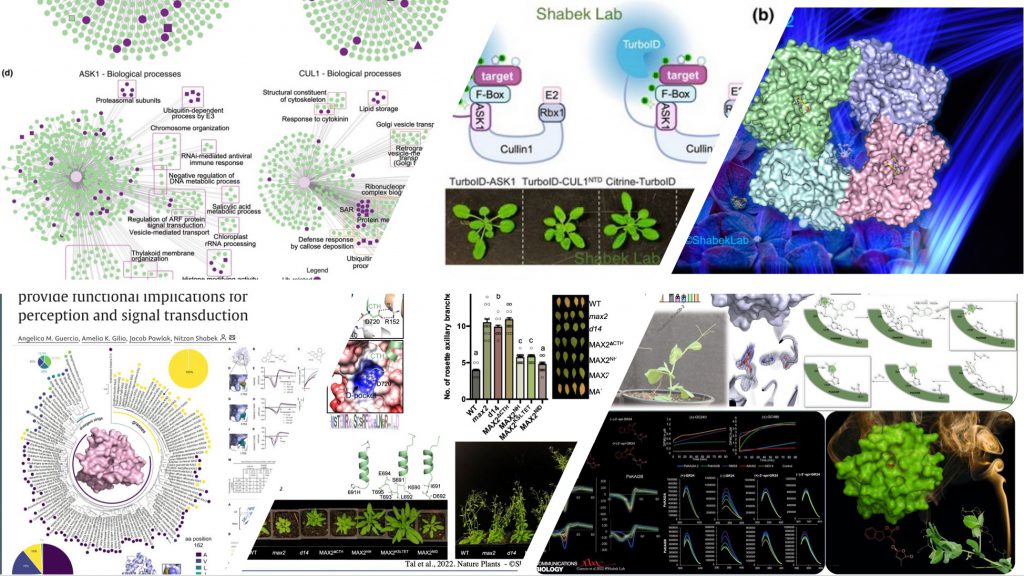 We are particularly interested in characterizing signal transduction pathways that are triggered by environmental stimuli. What are the precise biochemical adaptations, and how do they render a specific biomolecular sensor to be promptly recognized by yet another regulatory protein (e.g., ubiquitin ligase, transcription factor etc.)? All these questions remain to be answered for many biological pathways. In our lab we address these fundamental challenges by leveraging structural biology (X-ray crystallography and Cryo-EM) approaches in combination with biochemistry, molecular & cellular biology, plant biology, proteomics, and metabolomics methods.
We are particularly interested in characterizing signal transduction pathways that are triggered by environmental stimuli. What are the precise biochemical adaptations, and how do they render a specific biomolecular sensor to be promptly recognized by yet another regulatory protein (e.g., ubiquitin ligase, transcription factor etc.)? All these questions remain to be answered for many biological pathways. In our lab we address these fundamental challenges by leveraging structural biology (X-ray crystallography and Cryo-EM) approaches in combination with biochemistry, molecular & cellular biology, plant biology, proteomics, and metabolomics methods.
Read more about our main avenues of research (click on the links below):
Plant hormone signaling
Light signaling pathways
Ubiquitin biology
Applied research and biotechnology
The research in the Shabek Lab is largely funded and supported by:
The National Science Foundation (NSF-MCB EAGER, NSF-MCB CAREER, and NSF-IOS Awards)
The Department of Energy (DOE) – BER
OerthBio LLC (Bayer & Arvinas)
UC Davis (Innovation and Creativity Award and Academic Senate Large Research Grants)
UC Davis Environmental Health Sciences Core Center (EHSCC) Seed Grant
Students Fellowships include:
NIH (T32 Training Grant)
The National Institute of Food and Agriculture (NIFA), USDA-NIFA
NSF – Bioindustrial Engineering for a Sustainable Tomorrow (BEST training grant)
BARD, US-Israel Binational Agricultural Research and Development Fund
The Green Initiative Fellowship (TGIF)
Provost Undergrad Research Fellowships (PUFs)
Elsie Taylor Stocking Fellowship
Simon Chan Memorial Fellowship

